Gene regulatory programs driving metabolic maturation of human pluripotent stem cell derived β-cells
Contact PI: Han Zhu, PhD, University of Colorado Anschutz Medical Campus (R03 DK138495)
Start Date: November 15, 2023
*Moved from CHIB to CMAD on December 1, 2024
NIH HIRN Gateway Investigator Award Recipient
Abstract
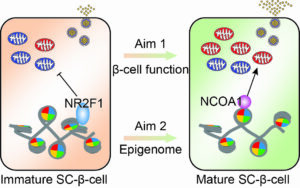
Type 1 diabetes (T1D) is characterized by autoimmune destruction of insulin-producing β-cells in the pancreatic islets. Several theories have been proposed to explain T1D pathogenesis including aberrant immune cell activity as well as increased β-cell sensitivity to inflammatory and metabolic stressors. It has become increasingly apparent that β-cells may not be passive immune cell targets but can contribute to the disease intrinsically. However, it remains unclear whether the risk of developing T1D is already introduced during β-cell formation. Postnatal β-cell maturation may be a T1D relevant β-cell developmental process, not only because it occurs during the peak incidence of T1D diagnosis but also because it determines whether an individual has an adequate functional β-cell pool to cope with environmental stressors. However, the mechanisms underlying human β-cell maturation have been understudied due to the absence of an appropriate human β-cell model. While human pluripotent stem cell (hPSC)-derived β-cells (SC-β-cells) hold immense potential for modeling T1D, current production methods do not permit proper functional maturation, hampering their utility in studying β-cell maturation. One of the hallmark features of mature β-cells is glucose-induced mitochondrial respiration, which is lacking in SC-β-cells. It has been suggested that environmental cues, such as nutrients and circadian rhythm, drive β-cell maturation and establish a mature β-cell state through gene expression regulation and epigenetic modifications. Our objective is to uncover gene regulatory mechanisms that underlie the acquisition of glucose-dependent mitochondrial function in human β-cells and assess the suitability of SC-β-cells as a model for studying β-cell maturation. This work will leverage the genomic and genetic methodologies that we have previously developed for SC-β-cells, including a platform for genome-wide functional perturbation screening, single-cell genomics assays, and a computational tool to infer gene regulatory networks. Expanding on our preliminary studies, we propose to investigate and modify key transcriptional programs governing SC-β-cell mitochondrial activity and induce maturation in vitro (Aim 1). Mechanistically, we plan to investigate the correlation between changes in SC-β-cell function and the epigenome that occurs during the maturation process (Aim 2). It has been established that SC-β-cells exhibit improved function following engraftment into mice. Thus, in the second aim of our study, we will also investigate epigenetic alterations that occur during this in vivo maturation process. We anticipate that our in vitro manipulation of key transcriptional programs will induce similar epigenetic remodeling as observed in vivo. Together, our proposed study will advance the culture of SC-β-cells and demonstrate their utility as a model for investigating human β-cell maturation.