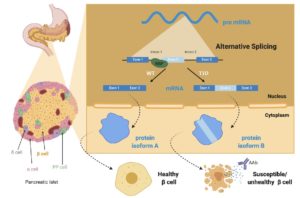
Alternative RNA splicing events contribute to the onset of islet dysfunction in T1D
Contact PI: Lori Sussel, PhD, University of Colorado Denver (U01 DK127505)
Charles Ansong, PhD, Investigator, Pacific Northwest National Laboratories (09/15/2020-10/31/2020)
Maggie (Pui Yu) Lam, PhD, co-Investigator, Pacific Northwest National Laboratories
Ernesto Nakayasu, PhD, co-Investigator, Pacific Northwest National Laboratory
Galya Orr, PhD, co-Investigator, Pacific Northwest National Laboratory
Shi Tujin, PhD, co-Investigator, Pacific Northwest National Laboratory
Start Date: September 15, 2020
End Date: December 2025
Abstract
Type 1 diabetes mellitus (T1D) is a chronic disease resulting from the autoimmune destruction of insulin- producing pancreatic b cells. Although T1D is primarily associated with a dysfunctional immune response, recent genome-wide association studies (GWAS) have determined that a large number of T1D associated genes are expressed in pancreatic b cells. Currently, there are many ongoing studies to identify the underlying molecular mechanisms that impact b cell dysfunction in T1D, with hopes that this knowledge can be used to develop novel biomarkers and preventive disease therapies. We hypothesize that altered RNA splicing events contribute to b cell dysfunction and potentially contribute to the formation of immunogenic protein isoforms to trigger T1D. Recently, our ability to perform genome wide RNA sequencing has revealed the complexity of the transcriptome and has provided significant insight into the transcriptomic modifications that occur during physiological and pathophysiological processes. In particular, there is a growing appreciation for the role of alternative splicing in many biological processes. The biological importance of alternative splicing has been further emphasized by the large number of human diseases caused by mutations that affect the splicing program. In the pancreas, a recent study showed considerable dysregulation of RNA binding proteins (RBPs) and aberrant mRNA splicing in islets derived from type 2 diabetic patients compared to healthy controls. While these studies have illustrated the existence of alternative splicing networks in the pancreas, and identified some of the enzymes involved in the regulation of alternative splicing, there has been relatively little demonstration of the functional consequences of alternative splicing events and how they may contribute to T1D pathogenesis. We propose studies to investigate the physiological relevance of alternative splicing events in b cells. Importantly, given the heterogeneous nature of b cell destruction in T1D, we will assess cell specific altered RNA splicing products at the single cell sequencing level, and in situ at a single cell and near single cell resolution to fully understand the potentially cell-specific functional consequences of altered spliced variants. Furthermore, we will identify potentially novel protein products that arise due to the alternative splicing events. To accomplish our goals, we propose the following specific aims: Aim 1. Use novel RNA sequencing technologies to identify conserved alternatively spliced transcripts in mouse and human T1D islet samples; Aim 2. Determine the localization of candidate RNA isoforms to individual human islet cells using fliFISH as a high accuracy smFISH approach that localizes individual transcripts with 20-30 nm resolution using STORM microscopy; Aim 3. Use advanced proteomics and innovative computational approaches to identify candidate novel protein products generated from alternative spliced transcripts in T1D b cells; and Aim 4. Determine the functional ramifications of altered splicing events in b cells by deleting a splicing factor important for regulating b cell function and survival.
Publications
- Human NKX2.2 influences islet endocrine cell fate choices through regulation of WNT pathway genes
- Challenges and Opportunities in State-of-the-Art Proteomics Analysis for Biomarker Development From Plasma Extracellular Vesicles
- Protein carbamylation and proteomics: from artifacts to elucidation of biological functions
- Optimizing Single-Cell Long-Read Sequencing for Enhanced Isoform Detection in Pancreatic Islets
- Insulin post-transcriptional regulation via PARP12-mediated ADP- ribosylation
- NKX2.2 and KLF4 cooperate to regulate α-cell identity
- Exploring new frontiers in type 1 diabetes through advanced mass-spectrometry-based molecular measurements
- The Chicken or the Egg Dilemma: Understanding the Interplay between the Immune System and the β Cell in Type 1 Diabetes
- Regulation of β-cell death by ADP-ribosylhydrolase ARH3 via lipid signaling in insulitis
- Reduction of chemokine CXCL9 expression by omega-3 fatty acids via ADP-ribosylhydrolase ARH3 in MIN6 insulin-producing cells
- A Composite Biomarker Signature of Type 1 Diabetes Risk Identified via Augmentation of Parallel Multi-Omics Data from a Small Cohort
- A fast and sensitive size-exclusion chromatography method for plasma extracellular vesicle proteomic analysis
- Protection of β cells against pro-inflammatory cytokine stress by the GDF15-ERBB2 signaling
- Modulation of insulin secretion by RBFOX2-mediated alternative splicing
- Systematic review of type 1 diabetes biomarkers reveals regulation in circulating proteins related to complement, lipid metabolism, and immune response
- Fatty acid-mediated signaling as a target for developing type 1 diabetes therapies
- Not the second fiddle: α cell development, identity and function in health and diabetes
- Plasma protein biomarkers predict the development of persistent autoantibodies and type 1 diabetes 6 months prior to the onset of autoimmunity
- Major β cell-specific functions of NKX2.2 are mediated via the NK2-specific domain
- Analysis of a macrophage carbamylated proteome reveals a function in post-translational modification crosstalk
- GDF15: a potential therapeutic target for type 1 diabetes
- From the dish to humans: A stem cell recipe for success
- Career Advancement for Women in Diabetes-Related Research: Developing and Retaining Female Talent
- Inherent Beta Cell Dysfunction Contributes to Autoimmune Susceptibility
- Islet Regeneration: Endogenous and Exogenous Approaches

