BioStructure & Function Team Leader
Contact PI: Ernesto Nakayasu, PhD, Pacific Northwest National Labortory
Start Date: December 2022
HIRN Catalyst Award Recipient
Abstract
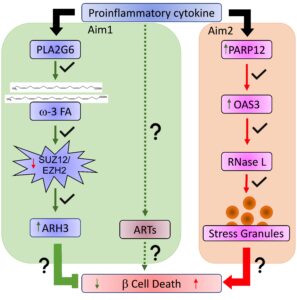
The goal of the project is to elucidate the signaling transduction mechanisms induced by omega-3 fatty acids that result in the protection of β cells from apoptosis. We proposed the following aims: in Aim 1, we will determine the ADP-ribosyltransferase responsible for triggering apoptosis and the ADP-ribosylated substrates involved in this process. We will perform a quantitative analysis of the ADP-ribosylated proteome followed by gene knockdown and western blot analysis to achieve this goal. Aim 2 will determine the dynamics of stress granules assembly and the impact of omega-3 fatty acids and PARP12 on it. We will also identify mRNAs sequestered in the stress granules and their translation regulation by PARP12.
Aim 1: To determine the mechanism of ADP-ribosylation mediated apoptosis and its regulation by omega-3 fatty acids. In this aim had proposed to determine the ADP-ribosyltransferase(s) ART(s) responsible for transducing the cytokine pro-apoptotic signaling by RNAi technology and luminescent apoptosis assay. Then to determine downstream substrates of ARTs, we would knockdown anti-apoptotic ADP-ribosylhydrolase ARH3 and identify potential apoptotic proteins that are over-ADP-ribosylated by proteomic analysis. Lastly, we would validate candidates by immunoprecipitation and anti-ADP-ribosylation Western blots in cells with both identified ART/s and ARH3 knockdown in combination with omega-3 and cytokine treatment.
Scope: We will perform proteomic analysis to determine the substrates of ARH3 only and we will not determine the substrates of ARTs identified to play a role in apoptosis. This should not compromise the final determination of the ADP-ribosylation-mediated β-cell apoptosis since they should be identified among the ARH3-regulated ADP-ribosylation sites. Overall, the proposed experiments should still determine the ADP-ribosylation factors and substrates involved in β-cell apoptosis.
Aim 2: Determine the role of PARP12 in stress-mediated translation halting. In this aim, we had proposed to investigate the roles of PARP12 in regulating protein translation. Our preliminary data indicate that PARP12 may participate in stress granules regulation by controlling factors such as IRGM1, IRF9, and OAS3. Therefore, we would test if the association of these proteins to stress granules depends on PARP12. We would also identify transcripts in that the translation is regulated by PARP12. In combination with Task 1 of Aim 1, the results of these experiments will show how PARP12 regulates protein translation and if this process is essential to mediate β cell protection by omega-3 fatty acids.
Scope: Our additional preliminary data indicates that insulin, GDF15 and PD-L1 transcripts are targeted to β cell stress granules. We have already validated that insulin mRNA targeting to stress granules is dependent of PARP12. Therefore, this reduces the need of identify new candidate transcripts that are targeted to stress granules by PARP12. Therefore, we will not perform RNA-seq of the ribosomal profiling that we had proposed. The ribosomal profiling will be followed by qPCR to quantify insulin, GDF15 and PD-L1 transcripts. Overall, the proposed experiments should still provide mechanistic insights on how PARP12 regulates the translational machinery of β cells during stress.