Integrative Analysis of Multiomic Signatures and Cellular Function in Human Pancreas Across Developmental Timeline at Single-cell Spatial Resolution
Contact PI: Marcela Brissova, PhD, Vanderbilt University (U01 DK135017)
Alejandro Caicedo, PhD, University of Miami
Jie Liu, PhD, University of Michigan
Stephen Parker, PhD, University of Michigan
Al Powers, MD, Vanderbilt University
Start Date: September 20, 2022
Abstract
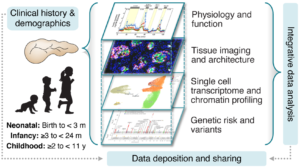
Studies of human pancreas development have begun to elucidate influences in the establishment of β cell mass and formation of islets, but genetic and environmental influences that manifest during postnatal pancreas development remain unknown. The first decade of life (termed the pediatric period for this proposal) is a dynamic time in pancreas development when two critcal processes occur: (1) β cell mass is established and (2) β cells and islets functionally mature. In addition, it is the time β cell-directed autoimmunity of type 1 diabetes (T1D) often begins. Thus, understanding the molecular and cellular processes that govern pediatric pancreas development and function is key to improving the diagnosis of children and adolescents with T1D and T2D and developing strategies to prevent, or treat the β cell dysfunction. While several ongoing initiatives including the Human Islet Research Network (HIRN) have been generating datasets from adult nondiabetic, T1D, and T2D donors, there is a major gap in deep molecular and tissue-level phenotyping of pancreata from the pediatric period. Furthermore, the contributions of vascular, immune, and other stromal cell populations and their β cell interactions, to human pediatric pancreas development are largely uncharacterized, despite their known influence on adult β cell function. Our proposal is based on our exciting single-cell multi-omic spatially-resolved pilot data that will allow us to map the context specificity of T1D and related trait GWAS signals in pancreas across cell type, age, sex, and developmental stage. Moreover, using living slice technology, we will be able to investigate cellular physiology and cell-cell communication in situ with high temporal resolution to provide an insight into processes that govern β cell maturation and establishment of healthy pancreatic architecture. The overlay of spatial, physiological, transcriptional, and chromatin data from the same organs will provide unprecedented access to define changes in molecular signatures, tissue architecture, and β cell maturation. This will not only complement phenotypic data collected from mostly adult donors in the Human Pancreas Analysis Program (HPAP), but will also generate data useful to several HIRN consortia and the broader research community. Our multidisciplinary research team with complementary expertise in pancreas and islet biology, in situ physiology, single cell genomics and epigenomics, image data analysis, statistical genetics, and machine learning devised tools and analyses to discover cell state dynamic changes across the first decade of life and define how these changes influence downstream biology from transcriptional regulation, to cellular spatial organization within the pancreas, and cellular function. If successful, these studies will provide new mechanistic insights about the functional maturation of human β cells during the critical pediatric life stages. This will likely influence the way we perceive T1D pathogenesis and lead to new therapies for diabetes and other pancreas diseases.
Publications
- CelLink: integrating single-cell multi-omics data with weak feature linkage and imbalanced cell populations
- Developing a general AI model for integrating diverse genomic modalities and comprehensive genomic knowledge
- Microvascular Homeostasis Is Compromised in Pancreatic Islets in a Mouse Model of β-Cell Loss and Low-Grade Inflammation
- A neural entero-pancreatic pathway that regulates insulin secretion and glucose tolerance
- The pathophysiology, presentation and classification of Type 1 diabetes
- Microvascular homeostasis is compromised in pancreatic islets in a mouse model of beta cell loss and low-grade inflammation
- Developing a general AI model for integrating diverse genomic modalities and comprehensive genomic knowledge
- SARS-CoV-2 Spike S1 subunit triggers pericyte and microvascular dysfunction in human pancreatic islets
- CNTools: A computational toolbox for cellular neighborhood analysis from multiplexed images
- Genetic risk converges on regulatory networks mediating early type 2 diabetes
- The human α cell in health and disease

