Epigenomic Engineering of Stem Cell-Derived Pancreatic Cells
Contact PI: Jeffrey Millman, Washington University School of Medicine in St. Louis
Start Date: September 2022
HIRN Catalyst Award Recipient
Abstract
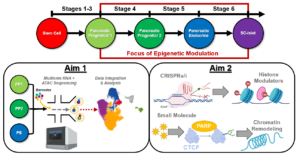
Epigenetics and chromatin accessibility have been shown to play an important role in cellular differentiation. While studies of epigenetics and chromatin accessibility have been broadly applied to developmental biology and stem cell biology, there has been no significant biological relevance applied within the context of stem cell-derived islet (SC-islet) development and maturation. SC-islets provide a revolutionary platform for both Type 1 diabetes (T1D) cell replacement therapy and in vitro modeling of diabetes pathogenesis. Using a temporal combination of small molecules and growth factors to recreate pancreatic development via activation and inhibition of developmental signaling pathways, we and others have determined the soluble and insoluble signals necessary to generate SC-islets capable of dynamic glucose-stimulated insulin secretion from human pluripotent stem cells (hPSCs). SC-islets can restore blood glucose homeostasis in diabetic mice and exhibit other features of primary human islets, such as expression of key transcription factors, heterogenous endocrine cell populations, and biphasic glucose-stimulated insulin secretion. However, two significant challenges limit the utility of SC-islets for cell replacement therapy and disease modeling: (1) SC-islets are functionally and transcriptionally immature compared to primary human islets and (2) existing protocols generate non-pancreatic off-target cell populations, specifically enterochromaffin cells (SC-ECs). The overarching goal of this proposal is to improve our understanding of the epigenetics of SC-islets and utilize this knowledge to influence cell fate choices during SC-islet differentiation. This will result in an improved SC-islet product that would be a powerful tool for studying stem cell biology, engineering islet tissue, modeling diabetes pathophysiology, and discovering novel T1D therapeutics. Thus, this proposal directly supports the goals set forth by HIRN, and more specifically the Consortium on Human Islet Biomimetics. Further, the application of novel multiomic sequencing to uncover fundamentally new insights related to SC-islets meets the needs of the Catalyst Award Program. To this end, the proposed research will generate new valuable insights through the following specific aims: (1) Identify cell-specific epigenomic regulators during SC-islet development, and (2) Develop optimized protocol to generate SC-islets through epigenetic modulation. Our approach utilizes the most advanced multiomic single cell sequencing available to simultaneously profile gene expression and open chromatin from the same cell. Thus, the proposed multimodal assessment of SC-islet development will provide the most detailed characterization of gene regulation and cell fate to date. This knowledge will not only provide an invaluable resource for the broader HIRN community, but also stimulate novel hypotheses to advance T1D research.
Publications

