Determining the Intrinsic and Environmental Signal Contributing to Early T1D Progression
Contact PI: Shuibing Chen, PhD, Weill Cornell Medical College (U01 DK127777)
Steven Parker, PhD, Investigator, University of Michigan
Chengyang Liu, PhD, Co-Investigator, University of Pennsylvania
Start Date: September 15, 2020
Abstract
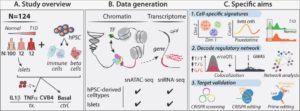
Type 1 diabetes (T1D) is caused by autoimmune response induced pancreatic β cell destruction. Both intrinsic (beta cell) and environmental (immune cell) signals play critical roles in pancreatic β cell dysfunction and death. Understanding the intrinsic and environmental network signature dynamics will facilitate dissecting the molecular mechanisms controlling T1D progression. Here, we build an interdisciplinary team with the expertise of diabetes computational and functional genomics, stem cell biology, and islet biology to systematically explore the intrinsic and environmental changes during T1D progression. In the preliminary studies, we performed single cell transcriptome (scRNA-seq) and chromatin (scATAC-seq) profiling of healthy, autoantibody positive (both non- hyperglycemia and hyperglycemia) patient islet samples, as well as a laboratory model that mimics T1D islet cell-specific signatures using human islets exposed to either cytokines or virus. In addition, we have created a platform to use isogenic human pluripotent stem cells (hPSCs)-derived pancreatic beta cells and macrophage- like cells to explore the biological function of diabetes associated genes or single nucleotide polymorphisms. Here, we will combine our expertise of diabetes computational and functional genomics and stem cell biology to systematically investigate the role of key intrinsic and environmental signal dynamics in T1D progression and establish the mechanistic network controlling pancreatic beta cell dysfunction. To achieve these goals, we propose three specific aims: Aim 1: Determine the cell-specific intrinsic and environmental signatures during T1D progression. Aim 2: Decode the cell-specific genetic regulatory network controlling T1D progression. Aim 3: Validate cells, genes, genetic variants, and intrinsic and environment signatures in T1D progression using an isogenic hPSC-based platform and primary T1D islets. Our key deliverables include: 1) a single-cell resolution multi-omic (scRNA-seq, scATAC-seq) map of healthy, T1D and cytokine- or CVB4 treated human islets; 2) the cell/context-specific molecular genetic (e/caQTL) network and hub signature of intrinsic and environmental signals in human pancreatic islets; 3) Isogenic hPSC- derived beta and immune cells with T1D-associated (hub) gene knockouts to validate individual or multiple genes. These results will be foundational for development of novel drugs and precision disease progression markers.
Publications
- Integrative single-cell multi-omics profiling of human pancreatic islets identifies T1D-associated genes and regulatory signals
- ChemPerturb-seq screen identifies a small molecule cocktail enhancing human beta cell survival after subcutaneous transplantation
- Single-cell transcriptomic profiling of human pancreatic islets reveals genes responsive to glucose exposure over 24 h
- Untangling the genetics of beta cell dysfunction and death in type 1 diabetes
- Checkpoint kinase 2 controls insulin secretion and glucose homeostasis
- An integrative single-cell multi-omics profiling of human pancreatic islets identifies T1D associated genes and regulatory signals
- Functional interrogation of twenty type 2 diabetes-associated genes using isogenic hESC-derived β-like cells
- Modeling islet enhancers using deep learning identifies candidate causal variants at loci associated with T2D and glycemic traits
- Human pluripotent-stem-cell-derived organoids for drug discovery and evaluation
- A multi-organoid platform identifies CIART as a key factor for SARS-CoV-2 infection
- A human iPSC-array-based GWAS identifies a virus susceptibility locus in the NDUFA4 gene and functional variants
- Cardiomyocytes recruit monocytes upon SARS-CoV-2 infection by secreting CCL2
- SARS-CoV-2 infection induces beta cell transdifferentiation
- An Immuno-Cardiac Model for Macrophage-Mediated Inflammation in COVID-19 Hearts
- Phenotypic technologies in stem cell biology

